Integrative Biological NMR

Group Leader

Group Leader
Our research focuses on the development of NMR methods and their applications, together with other biophysical techniques, in structural biology and to the biophysics of membranes, to characterize virulence factors or therapeutic targets associated with cancers or pathogen-host relationships, with a particular focus on mycobacterial infection.
An interdisciplinary research group elucidating molecular structure, dynamics, and interactions of biological processes using NMR, biophysical methods, and MD simulations.
The Integrative Biological NMR group applies its expertise and state-of-the-art equipment to pursue ambitious projects in structural biology with important implications in pharmacology. The study of structure, dynamics, and interactions of biomolecules are key to elucidating biochemical processes at atomic resolution. We continuously explore new avenues, such as the structure and function of complex membrane assemblies, and the influence of macromolecular dynamics on interactions, in the context of bacterial infection and cancer.
The relationship between host and pathogen is mediated by numerous biological processes. The molecular mechanisms by which the host detects and responds to infection by pathogens and by which pathogens circumvent host defences are of primary relevance to the development of novel therapeutic approaches.
– We are seeking to understand the defence mechanism of Mycobacterium tuberculosis against metal intoxication in human phagocytes, and to decipher the host’s response to pathogens mediated at a molecular level by a family of cell surface receptors. Upon infection by Mtb, macrophages attempt intoxication of the bacterium via an increase in the concentration of metals such as copper and zinc. In this context, the group of Dr. Olivier Neyrolles (IPBS) has identified three P1-type ATPases of Mtb (CtpC, CtpG and CtpV), each co-expressed with a small membrane-associated protein, PacL1, PacL2 and PacL3, respectively. While CtpC is required for optimal intracellular survival of Mtb at high zinc concentrations, PacL1 is required for zinc resistance in Mtb, it colocalises in foci with CtpC, and it stabilises CtpC efflux platforms at the plasma membrane. PacL1 therefore represents a novel type of metallochaperone in the sense that it binds zinc and stabilises CtpC. We are studying the selectivity of these metallochaperones towards metals and the mechanism of metal transfer towards the associated transporter.
– C-type lectin receptors (CLRs) are host proteins that play key roles in the innate immune system. CLRs allow cells to recognise characteristic molecules of pathogens or damaged cells and to trigger signalling pathways that culminate in protective effects. A better understanding of CLR signalling at the molecular level may facilitate the development of therapeutic agents targeting these receptors. Limited structural data is available, however, shedding only some light on ligand recognition. We focus specifically on a set of transmembrane CLRs (Mincle, Dectin-1, Dectin-2 and DCIR) that play key roles in pathologies such as infection, by Mtb and other bacteria, cancers, and autoimmune diseases. These CLRs are representatives of diverse signalling mechanisms. We aim to provide a detailed explanation of the allosteric coupling between extracellular ligand binding and intracellular partner protein recruitment that results in immune cell response.
Team members
Research Scientists
Andrew Atkinson (CNRS)
Cécile Bon (University)
Georges Czaplicki (University)
Pascal Demange (CNRS)
Guillaume Ferré (University)
Isabelle Muller (University)
Valérie Réat (CNRS)
Olivier Saurel (CNRS)
Research Engineers
Nathalie Doncescu (CNRS)
Pascal Ramos (University)
Pauline Rouan
PhD Students
Louis Bénastre
Thomas Benetti
Maxime Noriega
Our research projects
Deciphering the architecture and function of bacterial membrane efflux platforms
Understanding immune cell signalling through C-type lectin receptors
Further collaborations
Ferré et al. (2023) Sodium is a negative allosteric regulator of the ghrelin receptor. Cell Rep
Schahl et al. (2022) Evidence for amylose inclusion complexes with multiple acyl chain lipids using solid-state NMR and theoretical approaches. Carbohydr Polym
Ferré et al. (2019) Structure and dynamics of G protein-coupled receptor-bound ghrelin reveal the critical role of the octanoyl chain. Proc Natl Acad Sci USA
Augenstreich et al. (2019) The conical shape of DIM lipids promotes Mycobacterium tuberculosis infection of macrophages. Proc Natl Acad Sci USA
Saurel et al. (2017) Local and global dynamics in Klebsiella pneumoniae outer membrane protein a in lipid bilayers probed at atomic resolution. J Am Chem Soc
Carel et al. (2017) Identification of specific posttranslational O-mycoloylations mediating protein targeting to the mycomembrane. Proc Natl Acad Sci USA
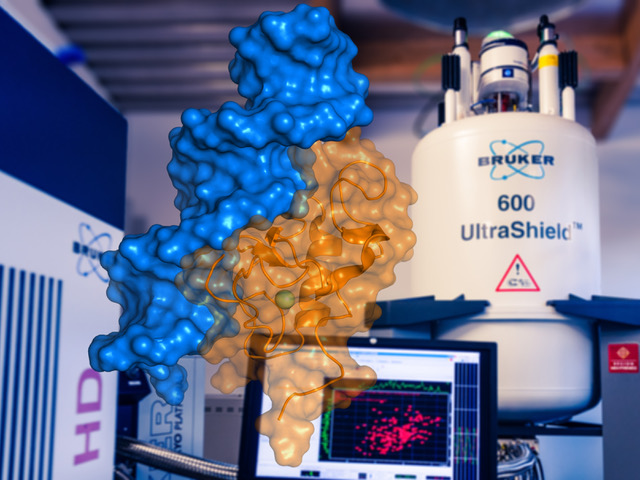
Macromolecular interactions by NMR: Solution structure of the THAP zinc finger of THAP1 in complex with its DNA target (PDB entry 2KO0). © Olivier Saurel
Institut de Pharmacologie et de Biologie Structurale (IPBS), Toulouse
Dr. Ahmed Amine Khamlichi Genetic Instability and Transcriptional Regulation
Dr. Lionel Mourey Structural Biophysics
Dr. Olivier Neyrolles Mycobacterial Interactions with Host Cells
Dr. Jérôme Nigou Immunomodulation by Mycobacterial Lipids and Glycoconjugates
G.CLIPS biotech
Dr. Rosie Dawaliby
Institut de Recherche en Santé Digestive (IRSD), Toulouse
Prof. Eric Oswald Pathogenesis and Commensalism of Enterobacteria
Laboratoire de Synthèse et Physico-Chimie de Molécules d’Intérêt Biologique (SPCMIB), Toulouse
Dr. Yves Génisson MoNALISA
Dr. Christian Lherbet SySMiC
Toulouse Biotechnology Institute (TBI), Toulouse
Dr. Didier Zerbib PHYGE – Integrated Physiology and Functional Genomics of
Microbial Systems
University of the Western Cape, South Africa
Prof. David J.R. Pugh Department of Biotechnology
Centre de Biologie Intégrative (CBI), Toulouse
Dr. Matthieu Chavent Multiscale Computational Immunology
Our laboratory is supported by grants from:
- Agence Nationale de la Recherche (ANR)
- Université de Toulouse III – Paul Sabatier
- Infrastructures en Biologie Santé et Agronomie (IBiSA)
- ERDF – REACT-EU
The complete list of our publications is available through Pubmed.
2024
Czaplicki, G., and Mazeres, S. (2024). Glophyt: a user-friendly, general-purpose program for nonlinear and multidimensional curve fitting via a hybrid stochastic and deterministic approach. Journal of Bioinformatics and Systems Biology 7, 219–226. https://doi.org/10.26502/jbsb.5107093.
Noriega, M., Corey, R.A., Haanappel, E., Demange, P., Czaplicki, G., Atkinson, R.A., and Chavent, M. (2024). Coarse-graining the recognition of a glycolipid by the C-type lectin mincle receptor. J. Phys. Chem. B. https://doi.org/10.1021/acs.jpcb.4c03242.
Menchon, G., Maveyraud, L., and Czaplicki, G. (2024). Molecular dynamics as a tool for virtual ligand screening. Methods Mol Biol 2714, 33–83. https://doi.org/10.1007/978-1-0716-3441-7_3.
Duncan, A.L., Gao, Y., Haanappel, E., Im, W., and Chavent, M. (2024). Recent advances in modeling membrane β-barrel proteins using molecular dynamics simulations: from their lipid environments to their assemblies. Methods Mol Biol 2778, 311–330. https://doi.org/10.1007/978-1-0716-3734-0_19.
Bories, P., Rima, J., Tranier, S., Marcoux, J., Grimoire, Y., Tomaszczyk, M., Launay, A., Fata, K., Marrakchi, H., Burlet-Schiltz, O., et al. (2024). HadBD dehydratase from Mycobacterium tuberculosis fatty acid synthase type II: A singular structure for a unique function. Protein Sci 33, e4964. https://doi.org/10.1002/pro.4964.
Baybekov, S., Llompart, P., Marcou, G., Gizzi, P., Galzi, J.-L., Ramos, P., Saurel, O., Bourban, C., Minoletti, C., and Varnek, A. (2024). Kinetic solubility: experimental and machine-learning modeling perspectives. Mol Inform 43, e202300216. https://doi.org/10.1002/minf.202300216.
Anazia, K., Koenekoop, L., Ferré, G., Petracco, E., Gutiérrez-de-Teran, H., and Eddy, M.T. (2024). Interaction networks within disease-associated GαS variants characterized by an integrative biophysical approach. J Biol Chem, 107497. https://doi.org/10.1016/j.jbc.2024.107497.
2023
Pritzlaff, A., Ferré, G., Dargassies, E., Williams, C.O., Gonzalez, D.D., and Eddy, M.T. (2023). Conserved protein-polymer interactions across structurally diverse polymers underlie alterations to protein thermal unfolding. ACS Cent Sci 9, 685–695. https://doi.org/10.1021/acscentsci.2c01522.
Kalakoutis, M., Pollock, R.D., Lazarus, N.R., Atkinson, R.A., George, M., Berber, O., Woledge, R.C., Ochala, J., and Harridge, S.D.R. (2023). Revisiting specific force loss in human permeabilised single skeletal muscle fibres obtained from older individuals. Am J Physiol Cell Physiol. https://doi.org/10.1152/ajpcell.00525.2022.
Ferré, G., Gomes, A.A.S., Louet, M., Damian, M., Bisch, P.M., Saurel, O., Floquet, N., Milon, A., and Banères, J.-L. (2023). Sodium is a negative allosteric regulator of the ghrelin receptor. Cell Rep 42, 112320. https://doi.org/10.1016/j.celrep.2023.112320.
Crosas-Molist, E., Graziani, V., Maiques, O., Pandya, P., Monger, J., Samain, R., George, S.L., Malik, S., Salise, J., Morales, V., et al. (2023). AMPK is a mechano-metabolic sensor linking cell adhesion and mitochondrial dynamics to Myosin-dependent cell migration. Nat Commun 14, 2740. https://doi.org/10.1038/s41467-023-38292-0.
Atkinson, R.A. (2023) NMR of proteins and nucleic acids. Nucl Magn Reson 49, 200-221. https://doi.org/10.1039/9781837672455
2022
Schahl, A., Lemassu, A., Jolibois, F., and Réat, V. (2022). Evidence for amylose inclusion complexes with multiple acyl chain lipids using solid-state NMR and theoretical approaches. Carbohydr Polym 276, 118749. https://doi.org/10.1016/j.carbpol.2021.118749.
Pritzlaff, A., Ferré, G., Mulry, E., Lin, L., Gopal Pour, N., Savin, D.A., Harris, M.E., and Eddy, M.T. (2022). Atomic-scale view of protein-PEG interactions that redirect the thermal unfolding pathway of PEGylated human galectin-3. Angew Chem Int Ed Engl 61, e202203784. https://doi.org/10.1002/anie.202203784.
Lauressergues, D., Ormancey, M., Guillotin, B., San Clemente, H., Camborde, L., Duboé, C., Tourneur, S., Charpentier, P., Barozet, A., Jauneau, A., et al. (2022). Characterization of plant microRNA-encoded peptides (miPEPs) reveals molecular mechanisms from the translation to activity and specificity. Cell Rep 38, 110339. https://doi.org/10.1016/j.celrep.2022.110339.
Jackson, V., Hermann, J., Tynan, C.J., Rolfe, D.J., Corey, R.A., Duncan, A.L., Noriega, M., Chu, A., Kalli, A.C., Jones, E.Y., et al. (2022). The guidance and adhesion protein FLRT2 dimerizes in cis via dual small-X3-small transmembrane motifs. Structure 30, 1354-1365.e5. https://doi.org/10.1016/j.str.2022.05.014.
Ferré, G., Anazia, K., Silva, L.O., Thakur, N., Ray, A.P., and Eddy, M.T. (2022). Global insights into the fine tuning of human A2AAR conformational dynamics in a ternary complex with an engineered G protein viewed by NMR. Cell Rep 41, 111844. https://doi.org/10.1016/j.celrep.2022.111844.
Demange, P., Joly, E., Marcoux, J., Zanon, P.R.A., Listunov, D., Rullière, P., Barthes, C., Noirot, C., Izquierdo, J.-B., Rozié, A., et al. (2022). SDR enzymes oxidize specific lipidic alkynylcarbinols into cytotoxic protein-reactive species. Elife 11, e73913. https://doi.org/10.7554/eLife.73913.
Chadelle, L., Liu, J., Choesmel-Cadamuro, V., Karginov, A.V., Froment, C., Burlet-Schiltz, O., Gandarillas, S., Barreira, Y., Segura, C., Van Den Berghe, L., et al. (2022). PKCθ-mediated serine/threonine phosphorylations of FAK govern adhesion and protrusion dynamics within the lamellipodia of migrating breast cancer cells. Cancer Lett 526, 112–130. https://doi.org/10.1016/j.canlet.2021.11.026.
Boudehen, Y.-M., Faucher, M., Maréchal, X., Miras, R., Rech, J., Rombouts, Y., Sénèque, O., Wallat, M., Demange, P., Bouet, J.-Y., et al. (2022). Mycobacterial resistance to zinc poisoning requires assembly of P-ATPase-containing membrane metal efflux platforms. Nat Commun 13, 4731. https://doi.org/10.1038/s41467-022-32085-7.
Atkinson, R.A. (2022) NMR of proteins and nucleic acids. Nucl Magn Reson 48, 249-270. https://doi.org/10.1039/9781839167690
2021
Young, J.D., Ma, M.T., Eykyn, T.R., Atkinson, R.A., Abbate, V., Cilibrizzi, A., Hider, R.C., and Blower, P.J. (2021). Dipeptide inhibitors of the prostate specific membrane antigen (PSMA): a comparison of urea and thiourea derivatives. Bioorganic & Medicinal Chemistry Letters, 128044. https://doi.org/10.1016/j.bmcl.2021.128044.
Schahl, A., Gerber, I.C., Réat, V., and Jolibois, F. (2021). Diversity of the hydrogen bond network and its impact on NMR Parameters of amylose B polymorph: a study using molecular dynamics and DFT calculations within periodic boundary conditions. J Phys Chem B 125, 158–168. https://doi.org/10.1021/acs.jpcb.0c08631.
Javaid, S., Zafar, H., Atia-Tul-Wahab, Gervais, V., Ramos, P., Muller, I., Milon, A., Atta-Ur-Rahman, and Choudhary, M.I. (2021). Identification of new lead molecules against anticancer drug target TFIIH subunit P8 using biophysical and molecular docking studies. Bioorg Chem 114, 105021. https://doi.org/10.1016/j.bioorg.2021.105021.
Hungnes, I.N., Al-Salemee, F., Gawne, P.J., Eykyn, T., Atkinson, R.A., Terry, S.Y.A., Clarke, F., Blower, P.J., Pringle, P.G., and Ma, M.T. (2021). One-step, kit-based radiopharmaceuticals for molecular SPECT imaging: a versatile diphosphine chelator for 99mTc radiolabelling of peptides. Dalton Trans 50, 16156–16165. https://doi.org/10.1039/d1dt03177e.
Baybekov, S., Marcou, G., Ramos, P., Saurel, O., Galzi, J.-L., and Varnek, A. (2021). DMSO solubility assessment for fragment-based screening. Molecules 26, 3950. https://doi.org/10.3390/molecules26133950.
Atkinson, R.A. (2021) NMR of proteins and nucleic acids. Nucl Magn Reson 47, 204-239. https://doi.org/10.1039/9781839164965
2020
Schahl, A., Réat, V., and Jolibois, F. (2020). Structures and NMR spectra of short amylose-lipid complexes. Insight using molecular dynamics and DFT quantum chemical calculations. Carbohydr Polym 235, 115846. https://doi.org/10.1016/j.carbpol.2020.115846.
Paganin-Gioanni, A., Rols, M.-P., Teissié, J., and Golzio, M. (2020). Cyclin B1 knockdown mediated by clinically approved pulsed electric fields siRNA delivery induces tumor regression in murine melanoma. Int J Pharm 573, 118732. https://doi.org/10.1016/j.ijpharm.2019.118732.
Nguyen, M.C., Saurel, O., Carivenc, C., Gavalda, S., Saitta, S., Tran, M.P., Milon, A., Chalut, C., Guilhot, C., Mourey, L., et al. (2020). Conformational flexibility of coenzyme A and its impact on the post-translational modification of acyl carrier proteins by 4’-phosphopantetheinyl transferases. FEBS J 287, 4729–4746. https://doi.org/10.1111/febs.15273.
Meilhoc, E., and Teissié, J. (2020). Electrotransformation of Saccharomyces cerevisiae. Methods Mol Biol 2050, 187–193. https://doi.org/10.1007/978-1-4939-9740-4_21.
Martinez, X., Chavent, M., and Baaden, M. (2020). Visualizing protein structures – tools and trends. Biochem Soc Trans 48, 499–506. https://doi.org/10.1042/BST20190621.
Manzo, G., Hind, C.K., Ferguson, P.M., Amison, R.T., Hodgson-Casson, A.C., Ciazynska, K.A., Weller, B.J., Clarke, M., Lam, C., Man, R.C.H., et al. (2020). A pleurocidin analogue with greater conformational flexibility, enhanced antimicrobial potency and in vivo therapeutic efficacy. Commun Biol 3, 697. https://doi.org/10.1038/s42003-020-01420-3.
Fontaine, C., Buscato, M., Vinel, A., Giton, F., Raymond-Letron, I., Kim, S.H., Katzenellenbogen, B.S., Katzenellenbogen, J.A., Gourdy, P., Milon, A., et al. (2020). The tissue-specific effects of different 17β-estradiol doses reveal the key sensitizing role of AF1 domain in ERα activity. Mol Cell Endocrinol 505, 110741. https://doi.org/10.1016/j.mce.2020.110741.
Fanelli, R., Berta, D., Földes, T., Rosta, E., Atkinson, R.A., Hofmann, H.-J., Shankland, K., and Cobb, A.J.A. (2020). Organocatalytic access to a cis-cyclopentyl-γ-amino acid: an intriguing model of selectivity and formation of a stable 10/12-helix from the corresponding γ/α-peptide. J. Am. Chem. Soc. 142, 1382–1393. https://doi.org/10.1021/jacs.9b10861.
Del Toro, D., Carrasquero-Ordaz, M.A., Chu, A., Ruff, T., Shahin, M., Jackson, V.A., Chavent, M., Berbeira-Santana, M., Seyit-Bremer, G., Brignani, S., et al. (2020). Structural basis of teneurin-latrophilin interaction in repulsive guidance of migrating neurons. Cell 180, 323-339.e19. https://doi.org/10.1016/j.cell.2019.12.014.
Dekoninck, K., Létoquart, J., Laguri, C., Demange, P., Bevernaegie, R., Simorre, J.-P., Dehu, O., Iorga, B.I., Elias, B., Cho, S.-H., et al. (2020). Defining the function of OmpA in the Rcs stress response. Elife 9. https://doi.org/10.7554/eLife.60861.
Augenstreich, J., Haanappel, E., Sayes, F., Simeone, R., Guillet, V., Mazeres, S., Chalut, C., Mourey, L., Brosch, R., Guilhot, C., et al. (2020). Phthiocerol dimycocerosates from Mycobacterium tuberculosis increase the membrane activity of bacterial effectors and host receptors. Front Cell Infect Microbiol 10, 420. https://doi.org/10.3389/fcimb.2020.00420.
Atkinson, R.A. (2020) NMR of proteins and nucleic acids. Nucl Magn Reson 46, 250-271. https://doi.org/10.1039/9781788010665
2019
Pasquet, L., Chabot, S., Bellard, E., Rols, M.-P., Teissié, J., and Golzio, M. (2019). Noninvasive gene electrotransfer in skin. Hum Gene Ther Methods 30, 17–22. https://doi.org/10.1089/hgtb.2018.051.
Pasquet, L., Bellard, E., Chabot, S., Markelc, B., Rols, M.-P., Teissié, J., and Golzio, M. (2019). Pre-clinical investigation of the synergy effect of interleukin-12 gene-electro-transfer during partially irreversible electropermeabilization against melanoma. J Immunother Cancer 7, 161. https://doi.org/10.1186/s40425-019-0638-5.
Martinez, X., Krone, M., Alharbi, N., Rose, A.S., Laramee, R.S., O’Donoghue, S., Baaden, M., and Chavent, M. (2019). Molecular graphics: bridging structural biologists and computer scientists. Structure 27, 1617–1623. https://doi.org/10.1016/j.str.2019.09.001.
Ladurantie, C., Coustets, M., Czaplicki, G., Demange, P., Mazères, S., Dauvillier, S., Teissié, J., Rols, M.-P., Milon, A., Ecochard, V., et al. (2019). A protein nanocontainer targeting epithelial cancers: rational engineering, biochemical characterization, drug loading and cell delivery. Nanoscale 11, 3248–3260. https://doi.org/10.1039/c8nr10249j.
Kotras, C., Fossépré, M., Roger, M., Gervais, V., Richeter, S., Gerbier, P., Ulrich, S., Surin, M., and Clément, S. (2019). A cationic tetraphenylethene as a light-up supramolecular probe for DNA G-quadruplexes. Front Chem 7, 493. https://doi.org/10.3389/fchem.2019.00493.
Hedger, G., Koldsø, H., Chavent, M., Siebold, C., Rohatgi, R., and Sansom, M.S.P. (2019). Cholesterol interaction sites on the transmembrane domain of the hedgehog signal transducer and class F G protein-coupled receptor smoothened. Structure 27, 549-559.e2. https://doi.org/10.1016/j.str.2018.11.003.
Guillet, V., Bordes, P., Bon, C., Marcoux, J., Gervais, V., Sala, A.J., Dos Reis, S., Slama, N., Mares-Mejía, I., Cirinesi, A.-M., et al. (2019). Structural insights into chaperone addiction of toxin-antitoxin systems. Nat Commun 10, 782. https://doi.org/10.1038/s41467-019-08747-4.
Galant, L., Delverdier, M., Lucas, M.-N., Raymond-Letron, I., Teissié, J., and Tamzali, Y. (2019). Calcium electroporation: the bioelectrochemical treatment of spontaneous equine skin tumors results in a local necrosis. Bioelectrochemistry 129, 251–258. https://doi.org/10.1016/j.bioelechem.2019.05.018.
Ferré, G., Louet, M., Saurel, O., Delort, B., Czaplicki, G., M’Kadmi, C., Damian, M., Renault, P., Cantel, S., Gavara, L., et al. (2019). Structure and dynamics of G protein-coupled receptor-bound ghrelin reveal the critical role of the octanoyl chain. Proc Natl Acad Sci U S A 116, 17525–17530. https://doi.org/10.1073/pnas.1905105116.
Ferré, G., Czaplicki, G., Demange, P., and Milon, A. (2019). Structure and dynamics of dynorphin peptide and its receptor. Vitam Horm 111, 17–47. https://doi.org/10.1016/bs.vh.2019.05.006.
Chabot, S., Bellard, E., Reynes, J.P., Tiraby, G., Teissié, J., and Golzio, M. (2019). Electrotransfer of CpG free plasmids enhances gene expression in skin. Bioelectrochemistry 130, 107343. https://doi.org/10.1016/j.bioelechem.2019.107343.
Augenstreich, J., Haanappel, E., Ferré, G., Czaplicki, G., Jolibois, F., Destainville, N., Guilhot, C., Milon, A., Astarie-Dequeker, C., and Chavent, M. (2019). The conical shape of DIM lipids promotes Mycobacterium tuberculosis infection of macrophages. Proc Natl Acad Sci U S A 116, 25649–25658. https://doi.org/10.1073/pnas.1910368116.
Abraham, M., Apostolov, R., Barnoud, J., Bauer, P., Blau, C., Bonvin, A.M.J.J., Chavent, M., Chodera, J., Čondić-Jurkić, K., Delemotte, L., et al. (2019). Sharing data from molecular simulations. J Chem Inf Model 59, 4093–4099. https://doi.org/10.1021/acs.jcim.9b00665.
2018
Menchon, G., Maveyraud, L., and Czaplicki, G. (2018). Molecular dynamics as a tool for virtual ligand screening. Methods Mol Biol 1762, 145–178. https://doi.org/10.1007/978-1-4939-7756-7_9.
Gervais, V., Muller, I., Mari, P.-O., Mourcet, A., Movellan, K.T., Ramos, P., Marcoux, J., Guillet, V., Javaid, S., Burlet-Schiltz, O., et al. (2018). Small molecule-based targeting of TTD-A dimerization to control TFIIH transcriptional activity represents a potential strategy for anticancer therapy. J Biol Chem 293, 14974–14988. https://doi.org/10.1074/jbc.RA118.003444.
Drożdż, W., Walczak, A., Bessin, Y., Gervais, V., Cao, X.-Y., Lehn, J.-M., Ulrich, S., and Stefankiewicz, A.R. (2018). Multivalent metallosupramolecular assemblies as effective DNA binding agents. Chemistry 24, 10802–10811. https://doi.org/10.1002/chem.201801552.
Drożdż, W., Bessin, Y., Gervais, V., Cao, X.-Y., Lehn, J.-M., Stefankiewicz, A.R., and Ulrich, S. (2018). Switching multivalent DNA complexation using metal-controlled cationic supramolecular self-assemblies. Chemistry 24, 1518–1521. https://doi.org/10.1002/chem.201705630.
Chavent, M., Karia, D., Kalli, A.C., Domański, J., Duncan, A.L., Hedger, G., Stansfeld, P.J., Seiradake, E., Jones, E.Y., and Sansom, M.S.P. (2018). Interactions of the EphA2 kinase domain with PIPs in membranes: implications for receptor function. Structure 26, 1025-1034.e2. https://doi.org/10.1016/j.str.2018.05.003.
Chavent, M., Duncan, A.L., Rassam, P., Birkholz, O., Hélie, J., Reddy, T., Beliaev, D., Hambly, B., Piehler, J., Kleanthous, C., et al. (2018). How nanoscale protein interactions determine the mesoscale dynamic organisation of bacterial outer membrane proteins. Nat Commun 9, 2846. https://doi.org/10.1038/s41467-018-05255-9.
2017
Saurel, O., Iordanov, I., Nars, G., Demange, P., Le Marchand, T., Andreas, L.B., Pintacuda, G., and Milon, A. (2017). Local and global dynamics in Klebsiella pneumoniae Outer membrane protein A in lipid bilayers probed at atomic resolution. J Am Chem Soc 139, 1590–1597. https://doi.org/10.1021/jacs.6b11565.
Duncan, A.L., Reddy, T., Koldsø, H., Hélie, J., Fowler, P.W., Chavent, M., and Sansom, M.S.P. (2017). Protein crowding and lipid complexity influence the nanoscale dynamic organization of ion channels in cell membranes. Sci Rep 7, 16647. https://doi.org/10.1038/s41598-017-16865-6.
Cukier, C.D., Tourdes, A., El-Mazouni, D., Guillet, V., Nomme, J., Mourey, L., Milon, A., Merdes, A., and Gervais, V. (2017). NMR secondary structure and interactions of recombinant human MOZART1 protein, a component of the gamma-tubulin complex. Protein Sci 26, 2240–2248. https://doi.org/10.1002/pro.3282.
Carel, C., Marcoux, J., Réat, V., Parra, J., Latgé, G., Laval, F., Demange, P., Burlet-Schiltz, O., Milon, A., Daffé, M., et al. (2017). Identification of specific posttranslational O-mycoloylations mediating protein targeting to the mycomembrane. Proc Natl Acad Sci U S A 114, 4231–4236. https://doi.org/10.1073/pnas.1617888114.
2016
Menchon, G., Bombarde, O., Trivedi, M., Négrel, A., Inard, C., Giudetti, B., Baltas, M., Milon, A., Modesti, M., Czaplicki, G., et al. (2016). Structure-based virtual ligand screening on the XRCC4/DNA Ligase IV interface. Sci Rep 6, 22878. https://doi.org/10.1038/srep22878.
Hemmann, J.L., Saurel, O., Ochsner, A.M., Stodden, B.K., Kiefer, P., Milon, A., and Vorholt, J.A. (2016). The one-carbon carrier methylofuran from Methylobacterium extorquens AM1 contains a large number of α- and γ-linked glutamic acid residues. J Biol Chem 291, 9042–9051. https://doi.org/10.1074/jbc.M116.714741.
Cukier, C.D., Maveyraud, L., Saurel, O., Guillet, V., Milon, A., and Gervais, V. (2016). The C-terminal region of the transcriptional regulator THAP11 forms a parallel coiled-coil domain involved in protein dimerization. J Struct Biol 194, 337–346. https://doi.org/10.1016/j.jsb.2016.03.010.
Brison, Y., Malbert, Y., Czaplicki, G., Mourey, L., Remaud-Simeon, M., and Tranier, S. (2016). Structural insights into the carbohydrate binding ability of an α-(1→2) branching sucrase from glycoside hydrolase family 70. J Biol Chem 291, 7527–7540. https://doi.org/10.1074/jbc.M115.688796.
2015
Sinnige, T., Houben, K., Pritisanac, I., Renault, M., Boelens, R., and Baldus, M. (2015). Insight into the conformational stability of membrane-embedded BamA using a combined solution and solid-state NMR approach. J Biomol NMR 61, 321–332. https://doi.org/10.1007/s10858-014-9891-6.
O’Connor, C., White, K.L., Doncescu, N., Didenko, T., Roth, B.L., Czaplicki, G., Stevens, R.C., Wüthrich, K., and Milon, A. (2015). NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor. Proc Natl Acad Sci U S A 112, 11852–11857. https://doi.org/10.1073/pnas.1510117112.
Berthoumieu, O., Nguyen, P.H., Castillo-Frias, M.P.D., Ferre, S., Tarus, B., Nasica-Labouze, J., Noël, S., Saurel, O., Rampon, C., Doig, A.J., et al. (2015). Combined experimental and simulation studies suggest a revised mode of action of the anti-Alzheimer disease drug NQ-Trp. Chemistry 21, 12657–12666. https://doi.org/10.1002/chem.201500888.
Bartolami, E., Bessin, Y., Gervais, V., Dumy, P., and Ulrich, S. (2015). Dynamic expression of DNA complexation with self-assembled biomolecular clusters. Angew Chem Int Ed Engl 54, 10183–10187. https://doi.org/10.1002/anie.201504047.
Former lab members have moved to fresh woods, and pastures new
- Dunkan Bégué | PhD student, Université de Nancy
- Evert Haanappel | CNRS Researcher, CBI, Toulouse
- Matthieu Chavent | CNRS Researcher, IPBS, Toulouse
- Adrien Schahl | Postdoctoral fellow, IPBS, Toulouse
- Virginie Gervais | CNRS Researcher I2BC, Saclay
- Guillaume Ferré | Lecturer, Université de Toulouse III – Paul Sabatier
- Nicolas Richet
- Marie Renault | Lecturer, Université de Toulouse III – Paul Sabatier
- Clément Carel | Masseur physiotherapist, Saint Orens de Gameville
- Guillaume Nars Doctor (endocrinology), CHU Strasbourg
- Parthasarathi Rath | Postdoctoral fellow, Biozentrum Basel, Switzerland
- Iordan Iordanov | Research associate, Semmelweis University, Budapest, Hungary
- Deborah Gater | Lecturer University, College London, United Kingdom
- Hélène Eury | Academic coordinator, Supexam, Rennes
- Olivier Cala | CNRS Research engineer, CRMN, Université de Lyon
- Léa Rougier | Teacher, College Elie Faure, Ste Foy La Grande
- Bruno Vincent | CNRS Engineer, Université de Strasbourg
- Corinne Hazan | General secretary, Artificial and Natural Intelligence Toulouse Institute
- Louic Vermeer | Data scientist, PGGM, Zeist, Netherlands
- Damien Bessiere | CNRS SAP engineer Toulouse
- Sébastien Campagne | INSERM Researcher, IECB, Bordeaux
- Masae Sugawara | Director of Paris Office Japan Science and Technology Agency, Paris
- Olivier Soubias | NIH Staff scientist, Bethesda, MD, USA

