Epigenetic Mechanisms in Cancer

Group Leader
Our laboratory is dedicated to studying the epigenetic mechanisms in cancer. We are specifically focused on understanding the functional role of arginine citrullination in transcriptional plasticity in cancer.
We aim to understand how the arginine citrullination regulates the transcription events and immune hijacking to provide better therapeutic approaches for cancer patients
Transcription regulation by RNA polymerase II is a fundamental process that is guided by tissue-specific transcription factors. These factors play a critical role in proliferation, cell identity, and function, and their dysregulation contributes to oncogenesis and drug resistance. Understanding the molecular mechanisms that regulate the recruitment of protein complexes to the transcription machinery is therefore essential for deciphering the fundamental processes underlying these pathological conditions.
Post-translational modifications of chromatin, including arginine citrullination, are crucial components of gene regulation mechanisms. Arginine citrullination is the conversion of arginine to the non-coded amino acid citrulline, catalyzed by a family of enzymes called peptidyl arginine deiminases (PADIs). We are deciphering the molecular mechanisms by which PADIs-mediated citrullination coordinates with transcriptional events at multiple levels and dictates transcriptional plasticity and immune regulation in cancer.
Given our expertise in the field of arginine citrullination and transcription regulation, we are intrigued to elucidate how arginine citrullination implicates the distinct stages of transcriptional events in cancer. Using advanced multidisciplinary approaches inclusive of single-cell omics, microscopy, proteomics, and the CRISPR-Cas9 method along with structural knowledge, we aim to gain a comprehensive understanding of the molecular mechanisms underlying this process. In combination, we are using functional approaches to translate this fundamental knowledge to provide better therapeutic strategies for cancer patients.
Team members
Research Scientist
Priyanka Sharma (Inserm)
Research Engineer
Cécile Meneur
Master Student
Molya Sajja
Villanueva-Cañas et al. (2024) Evolutionary analysis reveals the role of a non-catalytic domain of PADI2 in transcriptional regulation. iScience
Christophorou et al. (2023) Citrullination: new tricks for an old mod. Philos Trans R Soc Lond B Biol Sci
Beato & Sharma. (2020) Peptidyl arginine deiminase 2 (PADI2)-mediated arginine citrullination modulates transcription in cancer. Int J Mol Sci
Sharma et al. (2018) Arginine citrullination at the C-terminal domain controls RNA Polymerase II transcription. Mol Cell
Sharma et al. (2012) Citrullination of histone H3 interferes with HP1-mediated transcriptional repression. PLOS Genet
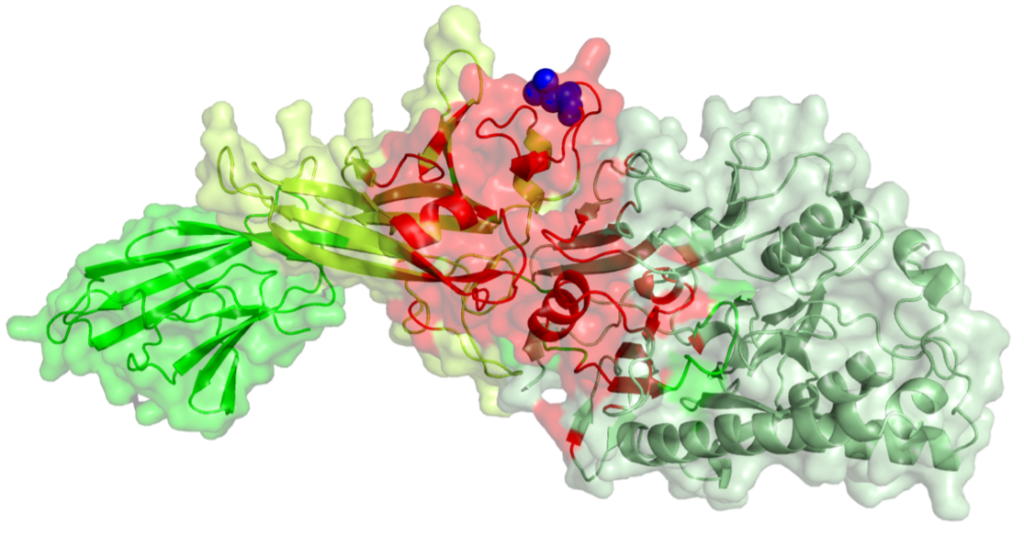
Surface representation of peptidyl arginine deiminase 2 with the consensus interface with the P-TEFb (positive transcription elongation factor b) complex derived from AlphaFold-Multimer and docking structural models highlighted in red and Leucine 162 in blue.
We are grateful to our collaborators, including:
• Prof. Juan Valcàrcel Juarez, Centre for Genomic Regulation (CRG), Barcelona, Spain
• Prof. Xavier Salvatella, Institute for Biomedical Research (IRB), Barcelona, Spain
• Dr. Daan Noordermeer, Institute for Integrative Biology of the Cell (I2BC), Université Paris Saclay, Paris, France
• Dr. Narcis Fernandez-Fuentes, Aberystwyth University, United Kingdom
• Prof. Baldomero Oliva, Department of Medicine and Life Sciences, University Pompeu Fabra, Barcelona, Spain
Funding

- Villanueva-Cañas, J.L., Fernandez-Fuentes, N., Saul, D., Kosinsky, R.L., Teyssier, C., Rogalska, M.E., Pérez, F.P., Oliva, B., Notredame, C., Beato, M. & Sharma, P. (2024). Evolutionary analysis reveals the role of a non-catalytic domain of PADI2 in transcriptional regulation, iScience . doi: https://doi.org/10.1016/j.isci.2024.109584.
- Christophorou MA, Sharma P, Zhang X. (2023). Citrullination: new tricks for an old mod. Philos Trans R Soc Lond B Biol Sci. 378:20220235.
- Beato M, Sharma P. (2021). Peptidyl Arginine Deiminase 2 (PADI2)-mediated Arginine Citrullination modulates Transcription in Cancer. Int J Mol Sci 21,1351
- Sharma, P., Lioutas, A., Fernandez-Fuentes, N., Quilez, J., Carbonell-Caballero, J., Wright, R. H. G., Di Vona, C., Le Dily, F., Schüller, R., Eick, D., Oliva, B., & Beato, M. (2019). Arginine citrullination at the C-terminal domain controls RNA Polymerase II transcription. Mol Cell, 73, 84-96
- Le Dily, F., Vidal, E., Cuartero, Y., Quilez, J., Nacht, A. S., Vicent, G. P., Carbonell-Caballero, J., Sharma, P., Villanueva-Cañas, J. L., Ferrari, R., De Llobet, L. I., Verde, G., Wright, R. H. G., & Beato, M. (2019). Hormone Control Regions mediate opposing steroid receptor-dependent genome organizations. Genome Res 29;29-39
- Kumar, A., Sharma, P., Gomar-Alba, M., Shcheprova, Z., Daulny, A., Sanmartín, T., Matucci, I., Funaya, C., Beato, M., & Mendoza, M. (2018). Daughter-Cell-specific modulation of nuclear pore complexes controls cell cycle entry during asymmetric division. Nat Cell Biol, 4, 432-442
- Nacht, A. S., Pohl, A., Zaurin, R., Soronellas, D., Quilez, J., Sharma, P., Wright, R. H., Beato, M., & Vicent, G. P. (2016). Hormone-induced repression of genes requires BRG1-mediated H1.2 deposition at target promoters. EMBO J, 35, 1822–1843
- Sharma, P., Azebi, S., England, P., Christensen, T., Møller-Larsen, A., Petersen, T., Batsché, E., & Muchardt, C. (2012). Citrullination of histone H3 interferes with HP1-mediated transcriptional repression. PLoS Genet, 8, e1002934
- Sharma, P., Garg, G., Kumar, A., Mohammad, F., Kumar, S. R., Tanwar, V. S., Sati, S., Sharma, A., Karthikeyan, G., Brahmachari, V., & Sengupta, S. (2014). Genome wide DNA methylation profiling for epigenetic alteration in coronary artery disease patients. Gene, 541, 31–40
- Sharma, P., Kumar, J., Garg, G., Kumar, A., Patowary, A., Karthikeyan, G., Ramakrishnan, L., Brahmachari, V., & Sengupta, S. (2008). Detection of altered global DNA methylation in coronary artery disease patients. DNA Cell Biol, 27, 357–365
- Sharma, P., Senthilkumar, R. D., Brahmachari, V., Sundaramoorthy, E., Mahajan, A., Sharma, A., & Sengupta, S. (2006). Mining literature for a comprehensive pathway analysis: a case study for retrieval of homocysteine related genes for genetic and epigenetic studies. Lipids Health Dis, 5, 1
- Halder, R., Halder, K., Sharma, P., Garg, G., Sengupta, S., & Chowdhury, S. (2010). Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol bioSystems, 6, 2439–2447
- Kumar, J., Das, S. K., Sharma, P., Karthikeyan, G., Ramakrishnan, L., & Sengupta, S. (2005). Homocysteine levels are associated with MTHFR A1298C polymorphism in Indian population. J Hum Genet, 50, 655–663

