Genetic Instability and Transcriptional Regulation

Group Leader
The aim of the team is to understand the transcriptional and epigenetic mechanisms that control the immunoglobulin heavy chain locus along B-cell development, and the role of the RAG complex and AID in B-cell acute lymphoblastic leukemia.
The major goal of our team is to decipher the molecular mechanisms underlying antibody diversity and B-cell cancers.
By using a combination of genetic, functional and mechanistic analyses, we investigate the molecular mechanisms that control antibody diversity and the role of specific enzymes in B-cell acute lymphoblastic leukemia.
Developing B lymphocytes have a remarkable capacity to somatically alter their genome through V(D)J recombination at the variable regions of their loci. This process is catalyzed by the RAG1/2 complex. Mature B cells have the unique ability to undergo an additional recombination process called class switch recombination (CSR), initiated by the enzyme AID (Activation-Induced cytidine Deaminase), which targets highly repetitive sequences within the constant genes of the immunoglobulin heavy chain (IgH) locus.
Both V(D)J recombination and CSR are controlled by distant regulatory elements on the chromosome. These include promoters, enhancers and insulators, which engage in long-range interactions in a developmental stage-specific manner.
Our aim is to understand the transcriptional and epigenetic mechanisms that control these interactions in vivo and how the cross-talk between IgH regulatory elements regulates IgH locus expression along B-cell development.
B-cell acute lymphoblastic leukemia (B-ALL) is the most frequent pediatric cancer. This neoplasm of early B cells is a multi-step disease featuring diverse genetic alterations. We have previously generated a mouse model which mimics the multistep leukemogenesis process of human B-ALL.
By using this model, our objective is to understand the role of the RAG complex and AID in the onset of B-ALL.
Team members
Research Scientist
Ahmed Amine Khamlichi (CNRS)
Research Engineers
Audrey Dauba (University)
PhD student
Kawtar Hanefioui
Our research projects
Transcriptional and epigenetic regulation of antibody diversification
Molecular mechanisms underlying B-cell acute lymphoblastic leukemia
Oudinet et al. (2022) Switch Tandem Repeats Influence the Choice of the Alternative End-Joining Pathway in Immunoglobulin Class Switch Recombination. Front Immunol
Dauba et al. (2021) Interleukin 7 regulates switch transcription in developing B cells. Cell Mol Immunol
Oudinet et al. (2020) Recombination may occur in the absence of transcription in the immunoglobulin heavy chain recombination centre. Nucleic Acids Res
Santos et al. (2019) Two modes of cis-activation of switch transcription by the IgH superenhancer. Proc Natl Acad Sci USA
Oudinet et al (2019) Developmental regulation of DNA cytosine methylation at the immunoglobulin heavy chain constant locus. PLoS Genet
Jamrog*, Chemin* et al. (2018) PAX5-ELN oncoprotein promotes multistep B-cell acute lymphoblastic leukemia in mice. Proc Natl Acad Sci USA
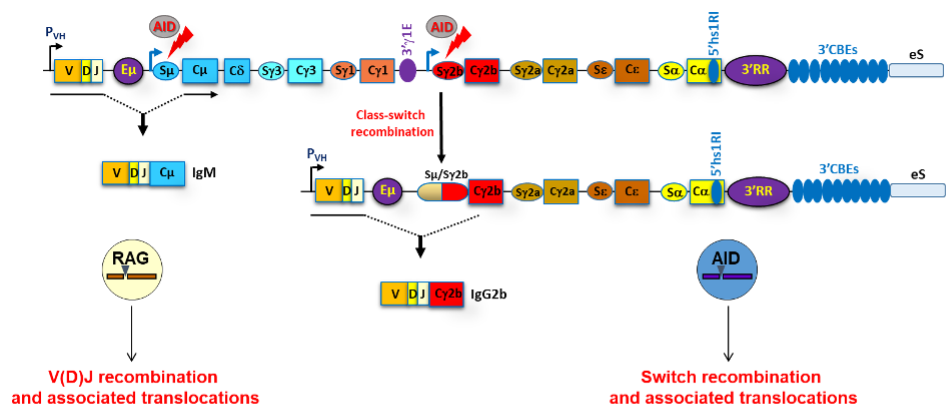
Scheme of a rearranged mouse IgH locus with its major regulatory elements. V(D)J recombination, CSR and associated chromosomal translocations are mediated by the RAG complex and AID respectively.
Collaborations
We are grateful to our past and present national and international collaborators:
- Frederick W Alt, Harvard Medical School, Boston, USA
- Michel Cogné, MOBIDIC INSERM, Rennes, France
- Eric Delabesse/Bastien Gerby, CRCT, INSERM, UPS, Toulouse, France
- Yves Denizot, CRIBL CNRS/INSERM, Limoges, France
- Robert Feil, IGMM CNRS, Montpellier, France
- Patricia J Gearhart, NIA / NIH, Baltimore, USA
- Zdenko Herceg, IARCancer, Lyon, France
- Amy L Kenter, University of Illinois College of Medicine, Chicago, USA
- Marc Le Bert, ENIM CNRS, Orléans, France
- Michael R Lieber, University of Southern California, Los Angeles, USA
Funding
- Agence Nationale de la Recherche
- Fondation ARC pour la recherche sur le cancer
- Institut National du Cancer
- Ligue contre le cancer
Prizes
June 2021 – Dr Chloé Oudinet is awarded the Toulouse Academy of sciences prize for her PhD work.
Press Releases
- Transcription versus recombination in the generation of antibody diversity
- In some instances, more is better…for the IgH super-enhancer
- DNA methylation: Better mark the locus before it’s too late…
- Pour une expression précise des gènes des anticorps, un commutateur temporel est nécessaire…
The complete list of our publications is available through Pubmed.
2024
- Dauba, A., Näser, E., Andrieux, D., Cogné, M., Denizot, Y., Khamlichi, A.A. The immunoglobulin heavy chain super enhancer controls class switch recombination in developing B cells. (2024). Sci Rep 14, 7370.
- Fregona, V., Bayet, M., Bouttier, M., Largeaud, L., Hamelle, C., Jamrog, L.A., Prade, N., Lagarde, S., Hebrard, S., Luquet, I., Mansat-De Mas, V., Nolla, M., Pasquet, M., Didier, C., Khamlichi, A.A., Broccardo, C., Delabesse, É., Mancini, S.J.C., Gerby, B. (2024). Stem cell-like reprogramming is required for leukemia-initiating activity in B-ALL. J Exp Med 221, e20230279.
2023
- Denis-Lagache, N, Oblet, C, Marchiol, T, Baylet, A, Têteau, O, Dalloul, I, Dalloul, Z, Zawil, L, Dézé, O, Cook-Moreau, J, Saintamand, A, Boutouil, H, Khamlichi, AA, Carrion, C, Péron, S, Le Noir, S, Laffleur, B, Cogné, M. (2023). Attempts to evaluate locus suicide recombination and its potential role in B cell negative selection in the mouse. Front Immunol 14, 1155906.
2022
- Oudinet, C., Zhang, X., Puget, N., Kyritsis, N., Leduc, C., Braikia, F.Z., Dauba, A., Alt, F.W., Khamlichi, A.A. (2022) Switch Tandem Repeats Influence the Choice of the Alternative End-Joining Pathway in Immunoglobulin Class Switch Recombination. Front Immunol 13, 870933
2021
- Dauba, A., Braikia, F.Z., Oudinet, C., Khamlichi, A.A. (2021) Interleukin 7 regulates switch transcription in developing B cells. Cell Mol Immunol 18, 776-778
- Dauba, A., Khamlichi, A.A. (2021) Long-Range Control of Class Switch Recombination by Transcriptional Regulatory Elements. Front Immunol 12, 738216
2020
- Oudinet, C., Braikia, F.Z., Dauba, A., Khamlichi, A.A. (2020) Mechanism and regulation of class switch recombination by IgH transcriptional control elements. Adv Immunol 147, 89-137
Oudinet, C., Braikia, F.Z., Dauba, A., Khamlichi, A.A. (2020) Recombination may occur in the absence of transcription in the immunoglobulin heavy chain recombination centre. Nucleic Acids Res 48, 3553-3566
2019
Oudinet, C., Braikia, F.Z., Dauba, A., Santos, J.M., Khamlichi, A.A. (2019) Developmental regulation of DNA cytosine methylation at the immunoglobulin heavy chain constant locus. PLoS Genet 15, e1007930
2018
- Khamlichi, A.A., Feil, R. (2018) Parallels between Mammalian Mechanisms of Monoallelic Gene Expression.Trends Genet 34, 954-971
- Jamrog, L., Chemin, G., Fregona, V., Coster, L., Pasquet, M., Oudinet, C., Rouquie, N., Prade, N., Lagarde, S., Cresson, C., Hebrard, S., Nguyen Huu, N.S., Bousquet, M., Quelen, C., Brousset, P., Mancini, S.J.C., Delabesse, E., Khamlichi, A.A., Gerby, B., Broccardo, C. (2018) PAX5-ELN oncoprotein promotes multistep B-cell acute lymphoblastic leukemia in mice. Proc Natl Acad Sci USA 115, 10357-10362115(41):10357-10362
- Santos, J.M., Braikia, F.Z., Oudinet, C., Haddad, D., Conte, C., Dauba, A., Khamlichi, A.A. (2018) Duplication of a germline promoter downstream of the IgH 3′ regulatory region impairs class switch recombination. Sci Rep 8, 9164
2017
- Braikia, F.Z., Oudinet, C., Haddad, D., Oruc, Z., Orlando, D., Dauba, A., Le Bert, M., Khamlichi, A.A. (2017) Inducible CTCF insulator delays the IgH 3′ regulatory region-mediated activation of germline promoters and alters class switching. Proc Natl Acad Sci USA 114, 6092-6097
2015
- Braikia, F.Z., Conte, C., Moutahir, M., Denizot, Y., Cogne, M., Khamlichi, A.A. (2015) Developmental Switch in the Transcriptional Activity of a Long-Range Regulatory Element. Mol Cell Biol 35, 3370-3380
- Puget, N., Leduc, C., Oruc, Z., Moutahir, M., Le Bert, M., Khamlichi, A.A. (2015) Complete cis Exclusion upon Duplication of the Emu Enhancer at the Immunoglobulin Heavy Chain Locus. Mol Cell Biol 35, 2231-2241
- Puget, N., Hirasawa, R., Hu, N.S., Laviolette-Malirat, N., Feil, R., Khamlichi, A.A. (2015) Insertion of an imprinted insulator into the IgH locus reveals developmentally regulated, transcription-dependent control of V(D)J recombination. Mol Cell Biol 35, 529-543
2014
- Braikia, F.Z., Chemin, G., Moutahir, M., Khamlichi, A.A. (2014) Quantification of V(D)J recombination by real-time quantitative PCR. Immunol Lett 162, 119-123
- Leduc, C., Chemin, G., Puget, N., Sawan, C., Moutahir, M., Herceg, Z., Khamlichi, A.A. (2014) Tissue-specific inactivation of HAT cofactor TRRAP reveals its essential role in B cells. Cell Cycle 13, 1583-1589
2011
- Haddad, D., Puget, N., Laviolette-Malirat, N., Conte, C., Khamlichi, A.A. (2011) Seeking sense of antisense switch transcripts. Transcription 2, 183-188
- Haddad, D., Oruc, Z., Puget, N., Laviolette-Malirat, N., Philippe, M., Carrion, C., Le Bert, M., Khamlichi, A.A. (2011) Sense transcription through the S region is essential for immunoglobulin class switch recombination. EMBO J 30, 1608-1620
2010
- Leduc, C., Haddad, D., Laviolette-Malirat, N., Nguyen Huu, N.S., Khamlichi, A.A. (2010) Combined deficiency of MSH2 and Smu region abolishes class switch recombination. Eur J Immunol 40, 2925-2931
- Zhang, T., Franklin, A., Boboila, C., McQuay, A., Gallagher, M.P., Manis, J.P., Khamlichi, A.A., Alt, F.W. (2010) Downstream class switching leads to IgE antibody production by B lymphocytes lacking IgM switch regions.Proc Natl Acad Sci USA 107, 3040-3045
- Haddad, D., Dougier, H.L., Laviolette, N., Puget, N., Khamlichi, A.A. (2010) Replacement of Imu-Cmu intron by NeoR gene alters Imu germ-line expression but has no effect on V(D)J recombination. Mol Immunol 47, 961-971
2009
- Rajagopal, D., Maul, R.W., Ghosh, A., Chakraborty, T., Khamlichi, A.A., Sen, R., Gearhart, P.J. (2009) Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med 206, 1237-1244
- Wang, L., Wuerffel, R., Feldman, S., Khamlichi, A.A., Kenter, A.L. (2009) S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med 206, 1817-1830
2007
- Oruc, Z., Boumediene, A., Le Bert, M., Khamlichi, A.A. (2007) Replacement of Igamma3 germ-line promoter by Igamma1 inhibits class-switch recombination to IgG3. Proc Natl Acad Sci USA 104, 20484-20489
- Tessier, J., Cuvillier, A., Glaudet, F., Khamlichi, A.A. (2007) Internalization and molecular interactions of human CD21 receptor. Mol Immunol 44, 2415-2425
- Huang, F.T., Yu, K., Balter, B.B., Selsing, E., Oruc, Z., Khamlichi, A.A., Hsieh, C.L., Lieber, M.R. (2007) Sequence dependence of chromosomal R-loops at the immunoglobulin heavy-chain Smu class switch region. Mol Cell Biol 27, 5921-5932
2006
Samara, M., Oruc, Z., Dougier, H.L., Essawi, T., Cogne, M., Khamlichi, A.A. (2006) Germ line transcription in mice bearing neor gene downstream of Igamma3 exon in the Ig heavy chain locus. Int Immunol 18, 581-589
We are indebted to former PhD students and post-doctoral fellows who are now continuing their career in the best labs, enterprises and institutions :
- Fatima-Zohra Braikia, post-doctoral fellow at the NIH / NIA, Baltimore, USA
- Guillaume Chemin, Assistant professor, Limoges University, France
- Dania Haddad, Research Assistant. Dasman Diabetes Institute, Kuwait City, Kuwait
- Claire Leduc, Marketing department. Syntivia, Toulouse, France
- Chloé Oudinet, Research Engineer, Evotec, Toulouse, France
- Joana M Santos, Group leader, CNRS, I2BC, Gif-sur-Yvette, France

