Deciphering & Drugging DNA Repair

Group Leader
The Deciphering & drugging DNA Repair (DDR) lab aims at deciphering how human cells respond to DNA damage to identify and validate new druggable complexes for cancer treatment. Our landmark discoveries include alternative End-Joining (2004), mechanisms controlling Ku association to DNA ends (2016, 2020, 2024), a new function for BRCA1 at centromeres (2014, 2021), the characterization of Ku interactions with DNA repair factors (2018, 2023, 2025) and the discovery of the mechanism of action of several bioactive small molecules (2017, 2021, 2022).
Through a multidisciplinary approach, we tackle an important problem: how DNA repair works and how can we modulate it through the use of small molecules.

DNA repair controls the outcome of several anticancer therapies. As such there is a huge interest for novel small molecules modulating DNA repair. In the DDR lab, we decipher how cells respond to DNA damage, more specifically to the most harmful type, DNA Double-Strand Breaks (DSBs). We use the resulting knowledge to design and perform targeted and phenotypical based screens to identify new modulators of specific aspects of the DNA damage response. To achieve our goals, we rely on high- and super-resolution imaging, molecular and cell biology, genomics, small molecules screening, chemical probes and on multiple collaborations with structural biologists and chemists. We recently implemented in the lab several complementary approaches to decipher how biologically active small molecules act (Bombarde et al. Mol Cancer Ther 2017; Bossaert, Pipier et al. 2021; Demange, Joly, Marcoux et al. eLife 2022).
The interplay between DNA repair mechanisms
Two principal DSB repair mechanisms co-exist in human cells: Non- Homologous End Joining (NHEJ) and Homologous Recombination (HR). Using a new method for imaging NHEJ proteins, we recently discovered the main mechanism antagonizing NHEJ proteins association to special DSBs, thereby allowing HR to proceed. We established that these mechanisms are druggable and that their inhibition triggers toxic DNA repair events in response to some anticancer agents (Britton et al. 2013 J Cell Biol; Chanut, Britton et al. 2016 Nat Commun; Britton, Chanut et al. 2020 Nucleic Acids Res).
How DNA repair proteins associate on damaged chromatin
We characterized how the repair factors APLF, XLF and the POLX associate with the NHEJ core protein Ku at DNA damage, thereby characterizing three new interaction interfaces at the functional and structural levels (Nemoz, Ropars, Frit et al. 2018 Nat Struct Mol Biol; Seif-El-Dahan, Kefala-Stavridi, Frit, Hardwick et al. 2023; Frit, Amin, Zahid, Barboule et al. 2025). We also revealed a novel role of DNA-PKcs in limiting Ku entry into chromatin (Bossaert, Moreno et al. 2024).
How stabilizing specific DNA structures triggers DNA damage
Small molecules stabilizing secondary DNA structures, such as G-quadruplexes (G4) ligands, trigger DNA damage (Zell et al. 2020 RSC Chem Biol). Through an unbiased genomic approach, we recently discovered that DNA topoisomerase 2 alpha is responsible for the production of DSB by several G4 ligands (Bossaert, Pipier et al. 2021 eLife), including CX-5461 a molecule undergoing clinical trials in oncology. We also contributed to develop novel ligands of another secondary structure, the Three-way Junction, and established that they also trigger DNA damage by a mechanism that we are currently investigating (Duskova 2019 J Med Chem; Duskova et al. 2020 J Am Chem Soc; Zell et al. Nucleic Acids Res 2021; Pipier et al. 2024).
Team members
Research Scientists
Nadia Barboule (CNRS)
Sébastien Britton (CNRS)
Patrick Calsou (Inserm)
Philippe Frit (CNRS)
Dennis Gomez (CNRS)
Léa Marie (University)
Florence Larminat (CNRS)
Marie-Jeanne Pillaire (Inserm)
Research Engineers
Antonio Peixoto (CNRS)
Emeline Pecourt
Carine Racca (CNRS)
PhD Students
Maëlle Caroff
Océane Dubourg
Bastien Dumais
Demange*, Joly*, Marcoux* et al. (2022) SDR enzymes oxidize specific lipidic alkynylcarbinols into cytotoxic protein-reactive species. eLife
Bossaert*, Pipier* et al. (2021) Transcription-associated topoisomerase 2alpha (TOP2A) activity is a major effector of cytotoxicity induced by G-quadruplex ligands. eLife
Racca et al. (2021) BRCA1 prevents R-loop-associated centromeric instability. Cell Death Dis
Britton*, Chanut* et al. (2020) ATM antagonizes NHEJ proteins assembly and DNA-ends synapsis at single-ended DNA double strand breaks. Nucleic Acids Res
Zell et al. (2021) Dual targeting of higher-order DNA structures by azacryptands induces DNA junction-mediated DNA damage in cancer cells. Nucleic Acids Res
Nemoz*, Ropars*, Frit* et al. (2018) XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining. Nat Struct Mol Biol
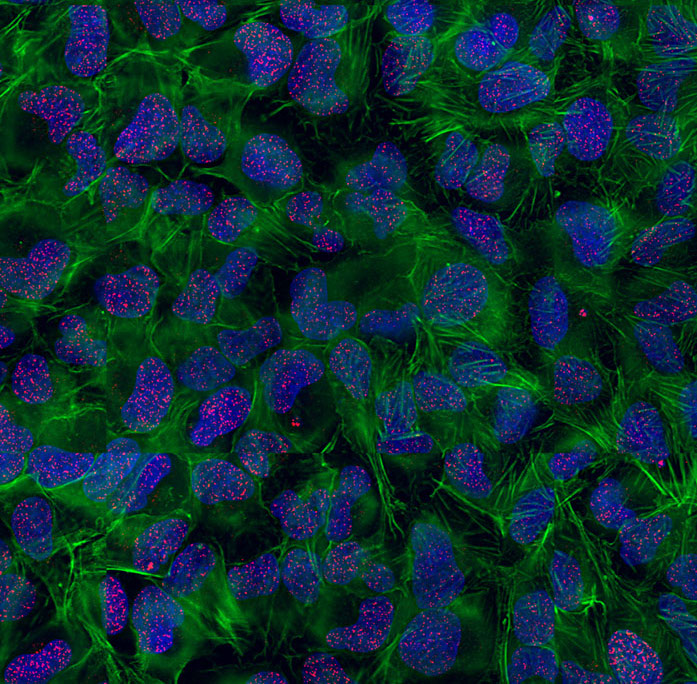
High-resolution imaging of DNA Double-Strand Breaks (DSBs) in human cells treated with ionizing radiations in presence of a DDR inhibitor. Red: DSBs visualized with gammaH2AX; Green: Actin cytoskeleton; Blue: DNA.
© Sébastien Britton
Collaborations
Local
- Françoise Benoit-Vical (LCC, UPR8241)
- Vania Bernardes-Génisson and Valérie Maraval (LCC, UPR 8241)
- Mamadou Daffé/Hedia Marrakchi (IPBS)
- Amine Khamlichi (IPBS)
- Yves Génisson (SPCMIB, UMR5068)
- Gaëlle Legube (CBI-LBCMCP)
- Lionel Mourey (IPBS)
- Stefania Millevoi (CRCT, UMR1037)
- Céline Noirot (Genotoul Bioinfo)
- Olivier Saurel/Andrew Atkinson (IPBS) and PICT platform
- Odile Schiltz (IPBS)
- Olivier Sordet (CRCT, UMR1037)
- Genotoul GeT-PlaGe
National
- Céline Gongora (IRCM, Montpellier)
- Yves-Jean Bignon and Nancy Uhrhammer (Centre Jean Perrin, Clermont-Ferrand)
- Isabelle Callebaut (IMPMC, Paris)
- Jean-Baptiste Charbonnier (I2BC, Saclay)
- Patrick Dallemagne (CERMN, Caen)
- Eric Defrancq (I2BM, Grenoble)
- Marc Delarue (Institut Pasteur, Paris)
- Daniele Fachinetti (Institut Curie, Paris)
- Claire Francastel (Paris Epigenetics, Paris)
- Anton Granzham (Institut Curie, Paris)
- Valérie Lamour (IGBMC, Strasbourg)
- Jean-Louis Mergny (IECB, Bordeaux)
- Mauro Modesti (CRCM, Marseille)
- David Monchaud (ICMUB, Dijon)
- Jean-François Riou (MNHN, Paris)
- Jean-Pierre de Villartay (Institut Imagine, Paris)
International
- Amanda Chaplin (University of Leicester)
- Steve Jackson (The Gurdon Institute, Cambridge, UK)
- Murray Junop (McMaster University, Canada)
- Susan Lees-Miller (University of Calgary, Canada)
- Michael Lieber (USC, Los Angeles, USA)
- Jo Loparo (Harvard Medical School, Boston, USA)
- Sankar Mitra (Houston Methodist Research Institute, USA)
- Eric van Dick (Luxembourg Institute of Health, Luxembourg)
Funding
- 2025 ANR “KIFMITO”, collaboration between F. Larminat, J. Basbous and S. Zinn-Justin.
- 2025 ANR “SEA-iDNA”, collaboration between D. Gomez, E. Defrancq, J. Dejeu and N Scaramozzino.
- 2025 Plan Cancer AVIESAN to S. Britton, collaboration with D. Monchaud, A. Granzhan, C. Gongora.
- 2024 TIRIS Scaling Up Science program “SDR-to-Lead” to S. Britton, collaboration with Y. Génisson
- 2024 Université Paul Sabatier TREMPLIN, to S. Britton, collaboration with Y. Génisson
- 2024 ANR “TOP2-G4”, to S. Britton, collaboration with V. Lamour and D. Monchaud
- 2023 Ligue contre le Cancer, Midi-Pyrénées grant to L. Marie.
- 2023 Fondation ARC, 4th year PhD fellowship to J. Chauvat.
- 2023 Fondation pour la Recherche Médicale, 4th year PhD fellowship to M. Bossaert.
- 2022 Fondation de France, Senior Post-doc fellowship to L. Marie.
- 2022 ANR “inJUNCTION”, collaboration between S.Britton, D. Monchaud, A. Granzhan, N. Chéron and L. Trantirek.
- 2022 ANR “MAG4”, to D.Gomez, collaboration with F. Benoit-Vical and V. Gervais.
- 2022 INCA PLBIO collaboration between M.-J. Pillaire, S. Manenti and C. Joffre.
- 2022 Roche ROADS program to A. Peixoto.
- 2022 Cancéropôle Grand-Sud-Ouest – Soutien à l’Emergence. Projet “CRIM”.
- 2022 Ligue Régionale contre le Cancer to S. Britton, collaboration with Y. Génisson.
- 2021 Prématuration Région Occitanie “Prostacure” to S. Britton, collaboration with Y. Génisson and V. Bernardes-Génisson/V.Maraval.
- 2021 ANR “iCARE”, collaboration between D. Gomez, E. Defrancq, M. Delarue, A. Granzhan, J.-L. Mergny, J.-F. Riou.
- 2020 ANR “BreakDance”, collaboration between P. Calsou, J.-B. Charbonnier, M. Delarue, M. Modesti.
- 2020 ANR “MELICENDRe”, collaboration between F. Larminat, C. Francastel and D. Fachinetti.
- 2019 Pre-Maturation grant from SATT-TTT to S. Britton, collaboration with Y. Génisson & R. Chauvin.
- 2019 Plan Cancer Aviesan “DDRi” to S. Britton, collaboration with P. Dallemagne, CERMN, Caen.
- 2019 Plan Cancer Aviesan “ANASTOMOSIS” to P.Calsou, collaboration with D. Monchaud, ICMUB, Dijon.
- 2019 Cancéropôle Grand-Sud-Ouest – Soutien à l’Emergence. Projet “KILR”.
- 2019 Cancéropôle Grand-Sud-Ouest – Soutien à l’Emergence. Projet “CX-Break”.
- 2018-2020 Labellisation Ligue Nationale Contre le Cancer “Cassures double-brin de l’ADN : mécanismes de réparation et connexions thérapeutiques en oncologie”.
- 2018 Programme Prématuration Région Occitanie “OPTIBREAK”, collaboration with Pascal demange, Lionel Mourey, Yves Génisson.
- 2017 ANR “NHEJ LIG4″, collaboration with M. Modesti and J.-B. Charbonnier”
- 2017 Fondation ARC. “Development of new anticancer agents”.
- 2017 ANR JCJC to S. Britton. “Drugging DNA repair complexes”.
- 2016 IDEX “Fishing out the intracellular target of novel antitumor pharmacophores bio-inspired from marine sponge” collaboration between S.Britton, Y. Génisson and R. Chauvin.
- 2016 ANR “G4-TopIPro” collaboration between D. Gomez, E. Defrancq and J.-F. Riou.
- 2016 Cancéropôle Grand Sud-Ouest – Soutien à l’Emergence (Cibles pharmacologiques de la Jaspine B) to S. Britton & P. Calsou.
- 2016 Cancéropôle Grand Sud-Ouest – Soutien à l’Emergence (G4POPSTAR, collaboration with G. Pratviel and S. Millevoi).
- 2016 Electricité de France – Conseil de Radioprotection.
- 2015 Electricité de France – Conseil de Radioprotection.
.
The complete list of our publications is available through Pubmed
Publications
* incidates (co-) corresponding authors ; # indicates equal contribution
2025
Le Bozec, B., Guitton-Sert, L., Collins, S., Finoux, A. L., Payrault, C., Guillou, E., Aguirrebengoa, M., Dougados, V., Jouffret, V., Frison, J., Carette, R., Rocher, V., Arnould, C., Guénolé, A., Lazar, I., Marnef, A., Frit, P., Calsou, P., Mangeat, T., Puget, N.*, Legube, G.* (2025) Circadian PERIOD proteins regulate TC-DSB repair through anchoring to the nuclear envelope. Mol Cell. (View in Molecular cell)
Bouvet, J.#, Mehalla, C.#, Vieira de Brito, M., Masson, L. B., Constant, P., Fabing, I., Preuilh, N., Listunov, D., Sinou, V., Conceição Ferreira Oliveira, M., Britton, S., Joly, E., Marrakchi, H.*, Maraval, V., Bernardes-Génisson, V.*, Ballereau, S.*, Génisson, Y.* (2025) Exploring the Chemical Reactivity of Triisopropylsilyl Dialkynylmethanol for the Synthesis of Dialkynylcarbinol‐Related Compounds. European J Med Chem. (View in European Journal of Medicinal Chemistry)
Vieira de Brito, M., Toublet, F.-X., Bossuat, M., Barbosa Davi, D. M., Kelly Almeida, D., de Sousa Fonseca, T., Miranda Nunes, F., Carlos de Mattos, M., Caroff, M., Britton, S., Ballereau, S., Maraval, V., Conceição Ferreira Oliveira, M.*, Génisson, Y.*, Bernardes-Génisson, V.* (2025) Exploring the Chemical Reactivity of Triisopropylsilyl Dialkynylmethanol for the Synthesis of Dialkynylcarbinol‐Related Compounds. European J Org Chem. (View in European Journal of Organic Chemistry)
Bossuat, M., Preuilh, N., Seigneur, P., Fabing, I., Pradel, C., Peixoto, A., Maraval, V., Bernardes-Génisson, V., Ballereau, S., Britton, S.*, Génisson, Y.* (2025) A plug and play approach to structural variations of bio-inspired anticancer lipidic phenyl dialkynylcarbinols. RSC Adv. (View in RSC Advances)
Frit, P.#, Amin, H.#, Zahid, S.#, Barboule, N.#, Hall, C., Matharu, G., Hardwick, S.W., Chauvat, J., Britton, S., Chirgadze, D.Y., Ropars, V., Charbonnier, J.-B., Calsou, P.*, Chaplin, A.K.* (2025) Structural and functional insights into the interaction between Ku70/80 and Pol X family polymerases in NHEJ. Nat Commun. (View in Nature communications).
2024
Bossaert, M. #, Moreno, A.T. #, Peixoto, A., Pillaire, M.-J., Chanut, P., Frit, P., Calsou, P.*, Loparo, J.J.*, Britton, S.* (2024) Identification of the main barriers to Ku accumulation in chromatin. Cell Rep. (View in Cell reports).
Pipier, A., Chetot, T., Kalamatianou, A., Martin, N., Caroff, M., Britton, S., Chéron, N., Trantírek, L., Granzhan, A.*, Monchaud, D.* (2024) Structural Optimization of Azacryptands for Targeting Three-Way DNA Junctions. Angew Chem Int Ed Engl. (View in Angewandte Chemie International Edition)
Boissieras, J., Bonnet, H., Susanto, M.F., Gomez, D., Defrancq, E., Granzhan, A.*, Dejeu, J.* (2024) iMab antibody binds single-stranded cytosine-rich sequences and unfolds DNA i-motifs. Nucleic Acids Res. (View in Nucleic Acids Research).
Gil Rosas, M., Centola, C., Torres, M., Mouguelar, V.S., David, A.P., Piga, E.J., Gomez, D., Calcaterra, N.B., Armas, P., Coux, G.* (2024) Sci Rep. (View in Scientific reports).
2023
Pinto, L.M.#, Pailas, A. #, Bondarchenko, M., Sharma, A.B., Neumann, K., Rizzo, A.J., Jeanty, C., Nicot, N., Racca, C., Graham, M.K., Naughton, C., Liu, Y., Chen, C.L., Meakin, P.J., Gilbert, N., Britton, S., Meeker, A.K., Heaphy, C.M., Larminat, F.*, Van Dyck, E.* (2023) DAXX promotes centromeric stability independently of ATRX by preventing the accumulation of R-loop-induced DNA double-stranded breaks. Nucleic Acids Res (View in Nucleic Acids Research)
Kefala Stavridi, A.#, Gontier, A. #, Morin, V. #, Frit, P. #, Ropars, V., Barboule, N., Racca, C., Jonchhe, S., Morten, M.J., Andreani, J., Rak, A., Legrand, P., Bourand-Plantefol, A., Hardwick, S.W., Chirgadze, D.Y., Davey, P., De Oliveira, T.M., Rothenberg, E., Britton, S.*, Calsou, P.*, Blundell, T.L.*, Varela, P.F.*, Chaplin, A.K.*, Charbonnier, J.B.* (2023) Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction. Nucleic Acids Res 51, 11732-11747 (View in Nucleic Acids Research)
Bossuat, M., Rulliere, P., Preuilh, N., Peixoto, A., Joly, E., Gomez, J.G., Bourkhis, M., Rodriguez, F., Goncalves, F., Fabing, I., Gaspard, H., Bernardes-Genisson, V., Maraval, V., Ballereau, S., Chauvin, R.*, Britton, S.*, Genisson, Y.* (2023) Phenyl dialkynylcarbinols, a Bioinspired Series of Synthetic Antitumor Acetylenic Lipids. J Med Chem 66, 13918-13945 (View in the Journal of Medicinal Chemistry)
Chebaiki, M., Delfourne, E., Tamhaev, R., Danoun, S., Rodriguez, F., Hoffmann, P., Grosjean, E., Goncalves, F., Azema-Despeyroux, J., Pal, A., Kordulakova, J., Preuilh, N., Britton, S., Constant, P., Marrakchi, H., Maveyraud, L.*, Mourey, L.*, Lherbet, C.* (2023) Discovery of new diaryl ether inhibitors against Mycobacterium tuberculosis targeting the minor portal of InhA. European journal of medicinal chemistry 259, 115646 (View in the European Journal of Medicinal Chemistry)
Seif-El-Dahan, M. #, Kefala-Stavridi, A. #, Frit, P. #, Hardwick, S.W. #, Chirgadze, D.Y., Maia De Oliviera, T., Andreani, J., Britton, S., Barboule, N., Bossaert, M., Pandurangan, A.P., Meek, K., Blundell, T.L., Ropars, V.*, Calsou, P.*, Charbonnier, J.B.*, Chaplin, A.K.* (2023) PAXX binding to the NHEJ machinery explains functional redundancy with XLF. Sci Adv 9, eadg2834 (View in Science Advances)
2022
Demange, P.#, Joly, E. #, Marcoux, J. #, Zanon, P.R.A., Listunov, D., Rulliere, P., Barthes, C., Noirot, C., Izquierdo, J.B., Rozie, A., Pradines, K., Hee, R., de Brito, M.V., Marcellin, M., Serre, R.F., Bouchez, O., Burlet-Schiltz, O., Oliveira, M.C.F., Ballereau, S., Bernardes-Genisson, V., Maraval, V., Calsou, P., Hacker, S.M., Genisson, Y.*, Chauvin, R.*, Britton, S.* (2022) SDR enzymes oxidize specific lipidic alkynylcarbinols into cytotoxic protein-reactive species. eLife 11 (View in eLife)
2021
Racca, C., Britton, S., Hedouin, S., Francastel, C., Calsou, P., Larminat, F.* (2021) BRCA1 prevents R-loop-associated centromeric instability. Cell death & disease 12, 896 (View in Cell Death & Disease)
Zell, J., Duskova, K., Chouh, L., Bossaert, M., Cheron, N., Granzhan, A., Britton, S.*, Monchaud, D.* (2021) Dual targeting of higher-order DNA structures by azacryptands induces DNA junction-mediated DNA damage in cancer cells. Nucleic Acids Res 49, 10275-10288 (View in Nucleic Acids Research)
Sharma, A.B., Erasimus, H., Pinto, L., Caron, M.C., Gopaul, D., Peterlini, T., Neumann, K., Nazarov, P.V., Fritah, S., Klink, B., Herold-Mende, C.C., Niclou, S.P., Pasero, P., Calsou, P., Masson, J.Y., Britton, S., Van Dyck, E. (2021) XAB2 promotes Ku eviction from single-ended DNA double-strand breaks independently of the ATM kinase. Nucleic Acids Res 49, 9906-9925 (View in Nucleic Acids Research)
Pipier, A., Devaux, A., Lavergne, T., Adrait, A., Coute, Y., Britton, S., Calsou, P., Riou, J.F., Defrancq, E., Gomez, D.* (2021) Constrained G4 structures unveil topology specificity of known and new G4 binding proteins. Scientific reports 11, 13469 (View in Scientific reports)
Bossaert, M.#, Pipier, A. #, Riou, J.F., Noirot, C., Nguyen, L.T., Serre, R.F., Bouchez, O., Defrancq, E., Calsou, P.*, Britton, S.*, Gomez, D.* (2021) Transcription-associated topoisomerase 2alpha (TOP2A) activity is a major effector of cytotoxicity induced by G-quadruplex ligands. eLife 10 (View in eLife)
2020
Zell, J., Rota Sperti, F., Britton, S.*, Monchaud, D.* (2021) DNA folds threaten genetic stability and can be leveraged for chemotherapy. RSC Chem Biol 2, 47-76 (View this review in RSC Chemical Biology)
Britton, S.*,#, Chanut, P. #, Delteil, C., Barboule, N., Frit, P., Calsou, P.* (2020) ATM antagonizes NHEJ proteins assembly and DNA-ends synapsis at single-ended DNA double strand breaks. Nucleic Acids Res 48, 9710-9723 (View in Nucleic Acids Research)
Janel-Bintz, R., Kuhn, L., Frit, P., Chicher, J., Wagner, J., Haracska, L., Hammann, P., Cordonnier, A.M.* (2020) Proteomic Analysis of DNA Synthesis on a Structured DNA Template in Human Cellular Extracts: Interplay Between NHEJ and Replication-Associated Proteins. Proteomics 20, e1900184 (View in Proteomics)
Duskova, K., Lejault, P., Benchimol, E., Guillot, R., Britton, S.*, Granzhan, A.*, Monchaud, D.* (2020) DNA Junction Ligands Trigger DNA Damage and Are Synthetic Lethal with DNA Repair Inhibitors in Cancer Cells. J Am Chem Soc 142, 424-435 (View in the Journal of the American Chemical Society)
2019
Cristini, A., Ricci, G., Britton, S., Salimbeni, S., Huang, S.N., Marinello, J., Calsou, P., Pommier, Y., Favre, G., Capranico, G., Gromak, N.*, Sordet, O.* (2019) Dual Processing of R-Loops and Topoisomerase I Induces Transcription-Dependent DNA Double-Strand Breaks. Cell reports 28, 3167-3181 e3166 (View in Cell Reports)
David, A.P., Pipier, A., Pascutti, F., Binolfi, A., Weiner, A.M.J., Challier, E., Heckel, S., Calsou, P., Gomez, D., Calcaterra, N.B., Armas, P.* (2019) CNBP controls transcription by unfolding DNA G-quadruplex structures. Nucleic Acids Res 47, 7901-7913 (View in Nucleic Acids Research)
Duskova, K., Lamarche, J., Amor, S., Caron, C., Queyriaux, N., Gaschard, M., Penouilh, M.J., de Robillard, G., Delmas, D., Devillers, C.H., Granzhan, A., Teulade-Fichou, M.P., Chavarot-Kerlidou, M., Therrien, B., Britton, S., Monchaud, D.* (2019) Identification of Three-Way DNA Junction Ligands through Screening of Chemical Libraries and Validation by Complementary in Vitro Assays. J Med Chem 62, 4456-4466 (View in the Journal of Medicinal Chemistry)
Pipier, A., De Rache, A., Modeste, C., Amrane, S., Mothes-Martin, E., Stigliani, J.L., Calsou, P., Mergny, J.L., Pratviel, G., Gomez, D. (2019) G-Quadruplex binding optimization by gold(iii) insertion into the center of a porphyrin. Dalton Trans 48, 6091-6099 (View in Dalton transactions)
Frit, P., Ropars, V., Modesti, M., Charbonnier, J.B.*, Calsou, P.* (2019) Plugged into the Ku-DNA hub: The NHEJ network. Progress in biophysics and molecular biology 147, 62-76 (View this review in Progress in Biophysics and Molecular Biology)
2018
Rozie, A., Santos, C., Fabing, I., Calsou, P., Britton, S.*, Genisson, Y., Ballereau, S.* (2018) Alkyne-Tagged Analogue of Jaspine B: New Tool for Identifying Jaspine B Mode of Action. Chembiochem 19, 2438-2442 (View in ChemBioChem)
Nemoz, C.#, Ropars, V. #, Frit, P. #, Gontier, A., Drevet, P., Yu, J., Guerois, R., Pitois, A., Comte, A., Delteil, C., Barboule, N., Legrand, P., Baconnais, S., Yin, Y., Tadi, S., Barbet-Massin, E., Berger, I., Le Cam, E., Modesti, M., Rothenberg, E., Calsou, P.*, Charbonnier, J.B.* (2018) XLF and APLF bind Ku80 at two remote sites to ensure DNA repair by non-homologous end joining. Nat Struct Mol Biol 25, 971-980 (View in Nature Structural & Molecular Biology)
2017
Bombarde, O., Larminat, F., Gomez, D., Frit, P., Racca, C., Gomes, B., Guilbaud, N., Calsou, P.* (2017) The DNA-Binding Polyamine Moiety in the Vectorized DNA Topoisomerase II Inhibitor F14512 Alters Reparability of the Consequent Enzyme-Linked DNA Double-Strand Breaks. Mol Cancer Ther 16, 2166-2177 (View in Molecular Cancer Therapeutics)
Gueddouda, N.M., Mendoza, O., Gomez, D., Bourdoncle, A.*, Mergny, J.L.* (2017) G-quadruplexes unfolding by RHAU helicase. Biochim Biophys Acta Gen Subj 1861, 1382-1388 (View in Biochimica et Biophysica Acta (BBA) – General Subjects)
2016
Chanut, P. #, Britton, S.*,#, Coates, J., Jackson, S.P.*, Calsou, P.* (2016) Coordinated nuclease activities counteract Ku at single-ended DNA double-strand breaks. Nat Commun 7, 12889 (View in Nature communications)
Hegde, M.L., Dutta, A., Yang, C., Mantha, A.K., Hegde, P.M., Pandey, A., Sengupta, S., Yu, Y., Calsou, P., Chen, D., Lees-Miller, S.P., Mitra, S. (2016) Scaffold attachment factor A (SAF-A) and Ku temporally regulate repair of radiation-induced clustered genome lesions. Oncotarget 7, 54430-54444 (View in Oncotarget)
Lamaa, A., Le Bras, M., Skuli, N., Britton, S., Frit, P., Calsou, P., Prats, H., Cammas, A., Millevoi, S.* (2016) A novel cytoprotective function for the DNA repair protein Ku in regulating p53 mRNA translation and function. EMBO Rep 17, 508-518 (View in EMBO reports)
Menchon, G., Bombarde, O., Trivedi, M., Negrel, A., Inard, C., Giudetti, B., Baltas, M., Milon, A., Modesti, M., Czaplicki, G., Calsou, P. (2016) Structure-Based Virtual Ligand Screening on the XRCC4/DNA Ligase IV Interface. Scientific reports 6, 22878 (View in Scientific reports)
Chabalier-Taste, C., Brichese, L., Racca, C., Canitrot, Y., Calsou, P., Larminat, F.* (2016) Polo-like kinase 1 mediates BRCA1 phosphorylation and recruitment at DNA double-strand breaks. Oncotarget 7, 2269-2283 (View in Oncotarget)
2015
Yuan, Y. #, Britton, S. #, Delteil, C., Coates, J., Jackson, S.P., Barboule, N., Frit, P., Calsou, P.* (2015) Single-stranded DNA oligomers stimulate error-prone alternative repair of DNA double-strand breaks through hijacking Ku protein. Nucleic Acids Res 43, 10264-10276 (View in Nucleic Acids Research)
Brown, J.S. #, Lukashchuk, N. #, Sczaniecka-Clift, M.†, Britton, S. †, le Sage, C., Calsou, P., Beli, P.*, Galanty, Y.*, Jackson, S.P.* (2015) Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites. Cell reports 11, 704-714 (View in Cell reports)
Sabater, L., Nicolau-Travers, M.L., De Rache, A., Prado, E., Dejeu, J., Bombarde, O., Lacroix, J., Calsou, P., Defrancq, E., Mergny, J.L., Gomez, D., Pratviel, G.* (2015) The nickel(II) complex of guanidinium phenyl porphyrin, a specific G-quadruplex ligand, targets telomeres and leads to POT1 mislocalization in culture cells. J Biol Inorg Chem 20, 729-738 (View in the Journal of Biological Inorganic Chemistry)
Douglas, P., Ye, R., Morrice, N., Britton, S., Trinkle-Mulcahy, L., Lees-Miller, S.P.* (2015) Phosphorylation of SAF-A/hnRNP-U Serine 59 by Polo-Like Kinase 1 Is Required for Mitosis. Mol Cell Biol 35, 2699-2713 (View in Molecular and Cellular Biology)
2014
Di Paolo, A., Racca, C., Calsou, P., Larminat, F.* (2014) Loss of BRCA1 impairs centromeric cohesion and triggers chromosomal instability. FASEB J 28, 5250-5261 (View in the FASEB Journal)
Britton, S. #, Dernoncourt, E. #, Delteil, C., Froment, C., Schiltz, O., Salles, B., Frit, P., Calsou, P.* (2014) DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res 42, 9047-9062 (View in Nucleic Acids Research)
Larrieu, D., Britton, S., Demir, M., Rodriguez, R.*, Jackson, S.P.* (2014) Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science 344, 527-532 (View in Science)
Frit, P., Barboule, N., Yuan, Y., Gomez, D., Calsou, P. (2014) Alternative end-joining pathway(s): bricolage at DNA breaks. DNA Repair (Amst) 17, 81-97 (View in DNA repair)
Abet, V., Mariani, A., Truscott, F.R., Britton, S., Rodriguez, R. (2014) Biased and unbiased strategies to identify biologically active small molecules. Bioorg Med Chem 22, 4474-4489 (View in Bioorganic & Medicinal Chemistry)
2013
Britton, S., Coates, J., Jackson, S.P.* (2013) A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J Cell Biol 202, 579-595 (View in the Journal of Cell Biology)
Cottarel, J., Frit, P., Bombarde, O., Salles, B., Negrel, A., Bernard, S., Jeggo, P.A., Lieber, M.R., Modesti, M., Calsou, P.* (2013) A noncatalytic function of the ligation complex during nonhomologous end joining. J Cell Biol 200, 173-186 (View in the Journal of Cell Biology)
Fallone, F., Britton, S., Nieto, L., Salles, B., Muller, C. (2013) ATR controls cellular adaptation to hypoxia through positive regulation of hypoxia-inducible factor 1 (HIF-1) expression. Oncogene 32, 4387-4396 (View in Oncogene)
2012
Rodriguez, R.#, Miller, K.M. #, Forment, J.V. #, Bradshaw, C.R., Nikan, M., Britton, S., Oelschlaegel, T., Xhemalce, B., Balasubramanian, S.*, Jackson, S.P.* (2012) Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol 8, 301-310 (View in Nature Chemical Biology)
2011
Cheng, Q., Barboule, N., Frit, P., Gomez, D., Bombarde, O., Couderc, B., Ren, G.S.*, Salles, B.*, Calsou, P.* (2011) Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res 39, 9605-9619 (View in Nucleic Acids Research)
Romera, C., Bombarde, O., Bonnet, R., Gomez, D., Dumy, P., Calsou, P., Gwan, J.F., Lin, J.H., Defrancq, E., Pratviel, G.* (2011) Improvement of porphyrins for G-quadruplex DNA targeting. Biochimie 93, 1310-1317 (View in Biochimie)
2010
Miller, K.M.#, Tjeertes, J.V. #, Coates, J., Legube, G., Polo, S.E., Britton, S., Jackson, S.P.* (2010) Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 17, 1144-1151 (View in Nature Structural & Molecular Biology)
Gomez, D., Guedin, A., Mergny, J.L., Salles, B., Riou, J.F., Teulade-Fichou, M.P., Calsou, P.* (2010) A G-quadruplex structure within the 5′-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res 38, 7187-7198 (View in Nucleic Acids Research)
Bombarde, O., Boby, C., Gomez, D., Frit, P., Giraud-Panis, M.J., Gilson, E., Salles, B., Calsou, P.* (2010) TRF2/RAP1 and DNA-PK mediate a double protection against joining at telomeric ends. EMBO J 29, 1573-1584 (View in the EMBO Journal)
Ye, J.#, Lenain, C. #, Bauwens, S., Rizzo, A., Saint-Leger, A., Poulet, A., Benarroch, D., Magdinier, F., Morere, J., Amiard, S., Verhoeyen, E., Britton, S., Calsou, P., Salles, B., Bizard, A., Nadal, M., Salvati, E., Sabatier, L., Wu, Y., Biroccio, A., Londono-Vallejo, A., Giraud-Panis, M.J., Gilson, E.* (2010) TRF2 and apollo cooperate with topoisomerase 2alpha to protect human telomeres from replicative damage. Cell 142, 230-242 (View in Cell)
2009
Britton, S., Froment, C., Frit, P., Monsarrat, B., Salles, B.*, Calsou, P. (2009) Cell nonhomologous end joining capacity controls SAF-A phosphorylation by DNA-PK in response to DNA double-strand breaks inducers. Cell Cycle 8, 3717-3722 (View in Cell cycle)
Britton, S., Frit, P., Biard, D., Salles, B.*, Calsou, P. (2009) ARTEMIS nuclease facilitates apoptotic chromatin cleavage. Cancer Res 69, 8120-8126 (View in Cancer research)
Wu, P.Y., Frit, P., Meesala, S., Dauvillier, S., Modesti, M., Andres, S.N., Huang, Y., Sekiguchi, J., Calsou, P., Salles, B., Junop, M.S. (2009) Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol 29, 3163-3172 (View in Molecular and Cellular Biology)
Britton, S., Salles, B., Calsou, P. (2008) c-MYC protein is degraded in response to UV irradiation. Cell Cycle 7, 63-70 (View in Cell cycle)

